We need more and it needs to be better
ML in the life sciences won't sing until the data improves
The JP Morgan Health Conference was this week and it was full of biotech and techbio types. There was a lot of talk about machine learning and how it might help those in medical need. DeepMind’s Isomorphic Labs announced two partnerships; insitro and Eikon presented at the conference itself. Recursion held a capstone event jointly with Nvidia, where Nvidia founder Jensen Huang openly wondered if we’d collected all the data we might need to tackle drug development and that it might just need the right hardware and processing. He also hilariously noted that life scientists are a gloomy lot, since we use words like “inhibit”. Some gloomy audience members whispered to themselves, “no, we do not have enough.” So do we? Pubmed has 36M publications. How much could we need?
We need more
Let’s consider a simple problem: ligands (chemicals) binding to protein targets. Such interactions are the basis of all small molecule medicines and folks have been trying to use computers to predict which pairs might bind for decades, using increasingly fancy methods. The most prominent of these efforts use rules-based approaches (think physics equations) or diffusion (start with the result you want, and show noisier and noisier examples to the computer to get it predict backwards). Generally speaking, large datasets win out over rules-based approaches (go read the Bitter Lesson for a history on this, I’ve written about it too). Unless ligand-binding is different from the other problems solved with large datasets - problems as diverse as image recognition, language translation, Go, and Atari games - and protein folding! - we’ll probably want a large ligand dataset. Is there one?
No.
There are a handful of large-ish small molecule/bioactivity datasets. Perhaps the best-curated one is bindingdb, which has maybe 1.3M interactions. You clean those up (deduplicate, remove contributions from inconsistent labs, p-hacking, etc) and you have about 400K. It took 300B tokens (words) to train GPT3 last year: that’s almost a million times more data than the best publicly-available ligand-target dataset.
GPT4, which is vastly better than GPT3, took 13T tokens.
So maybe we don’t clean up the data, hoping to make use of more examples. Maybe we use Pubchem, which has almost 300M bioactivities across different modalities, contributed by a lot of different labs. Could all those examples save us?
No.
It needs to be better
One problem with machine learning approaches is that they cheat. My favorite example comes from this paper, which examined object classification in images cobbled together from many sources. One source was a commercial equestrian photographer who used a watermark in each photo, and by using activation maps, the paper’s authors figured out the ML was not looking for the horse but looking for the watermark.
If you’re applying ML to your biological problem of interest and got the data from somebody else, how certain are you that there aren’t other watermarks hiding inside? Folks do cellular assays in plates all the time, but if you don’t randomize the positions of those treatments in the plates, the ML will just learn to predict well position (this is because cells on the outer edges have metabolic differences relative to those in the middle and those differences are very loud).
Are well positions and randomization strategies reported in the bioactivities we can get from Pubchem? They are not. You could train ML on such data, make strong predictions on your test-train split, but since it only learned well position and not biological activities, it will not generalize to new datasets correctly. Makes me wonder how this Kaggle competition is going, given the reported plate layout.
A great example of missing watermarks in the life sciences can be found in this paper: the original NIH ENCODE group checked the transcriptomes of a number of organs in human and mouse, and when they clustered these samples, they found that all the mouse organs looked more like each other than the corresponding organ in human (e.g., mouse heart looked more like mouse liver than mouse heart looked like human heart).
This is odd! Those organs do very different jobs and we’d expect hearts to look like hearts.
The missing watermark here was identifiable from the metadata from the DNA sequencing reads - each read identifies the instrument it was run on - and those mouse-mouse and human-human samples clustered not on mammalian species, but on the lab where they were prepared. These are high-dimensional datasets - like 20,000 dimensions - and light nudges across all of those due to sample processing can be stronger than the genuine organ type differences. With the right batch correction method, we find that, yes, hearts look like hearts and livers look like livers. You’d never figure that out without the metadata recording the sample handling, though. Even the ENCODE authors didn’t figure it out, and they’re the ones who generated the data.
Let’s collect more, better, data
So maybe the existing data aren’t sufficient. To solve these problems (even the ligand problem, which is much easier than cellular behaviors, which are much easier than cell-to-cell interactions, and don’t even get me started on organ systems or neuro), we’ll have to generate more data, and that data will have to be better. We have to creatively pre-empt potential watermarks with smart design, and we have to exhaustively record experimental metadata to mitigate other watermarks that elude our imagination. We have to check carefully for data leaks and kick the tires on these new collection approaches. And after all that, we’re going to have to collect at least a million times as much.
(At Leash, we’re doing this.)
Postscript
I started this piece referring to the 36M papers in Pubmed. Can those be useful in aggregate?
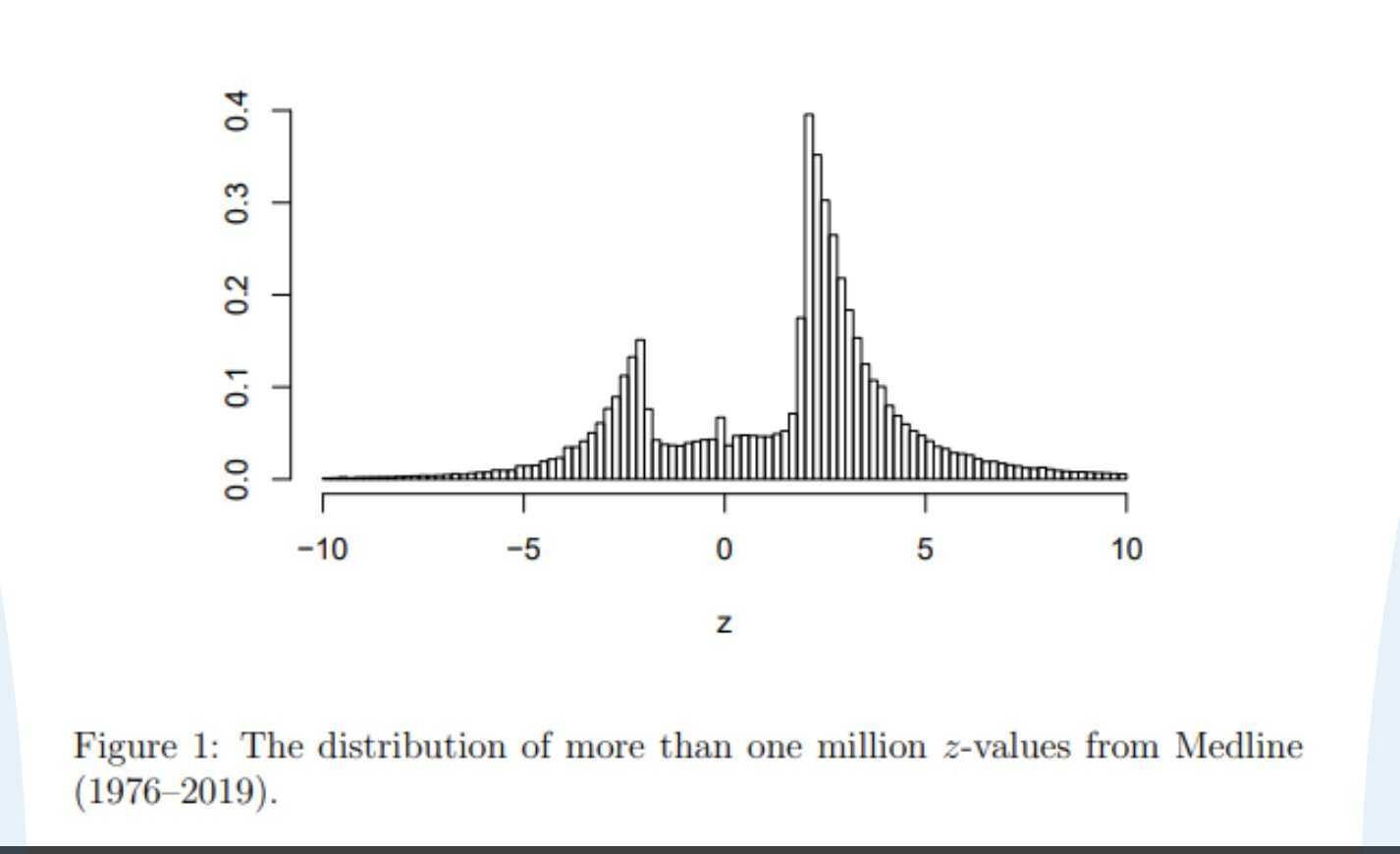
The very first post in Leash’s #general channel in Slack is the below figure from this paper. That hole in the middle? It’s where everything above a p-value of 0.05 should be. That plot should be a normal distribution! This means authors with nonsignificant results don’t publish their papers as often, and so we have a very incomplete picture.
No, we can’t rely on those papers in aggregate.