Automating the Drug Discovery Lab of the Future: Part 1
A series on automation, data generation, and analysis at Leash
-Our lab has moved out of Ian’s basement and is now fully operational at the Altitude Labs incubator. We’re now running the full workflow from soup to nuts.
It’s been just over a year since I joined Leash as Head of Operations and I’m way overdue to give an update. I thought it would be interesting to share my thoughts on automated laboratory data generation and what we’ve been up. I have a lot of thoughts, so I’ll be releasing this as a four part series:
Introduction
Data Acquisition
Data Analysis
Physical Automation
Part 1. Introduction
What We’ve Been Up To
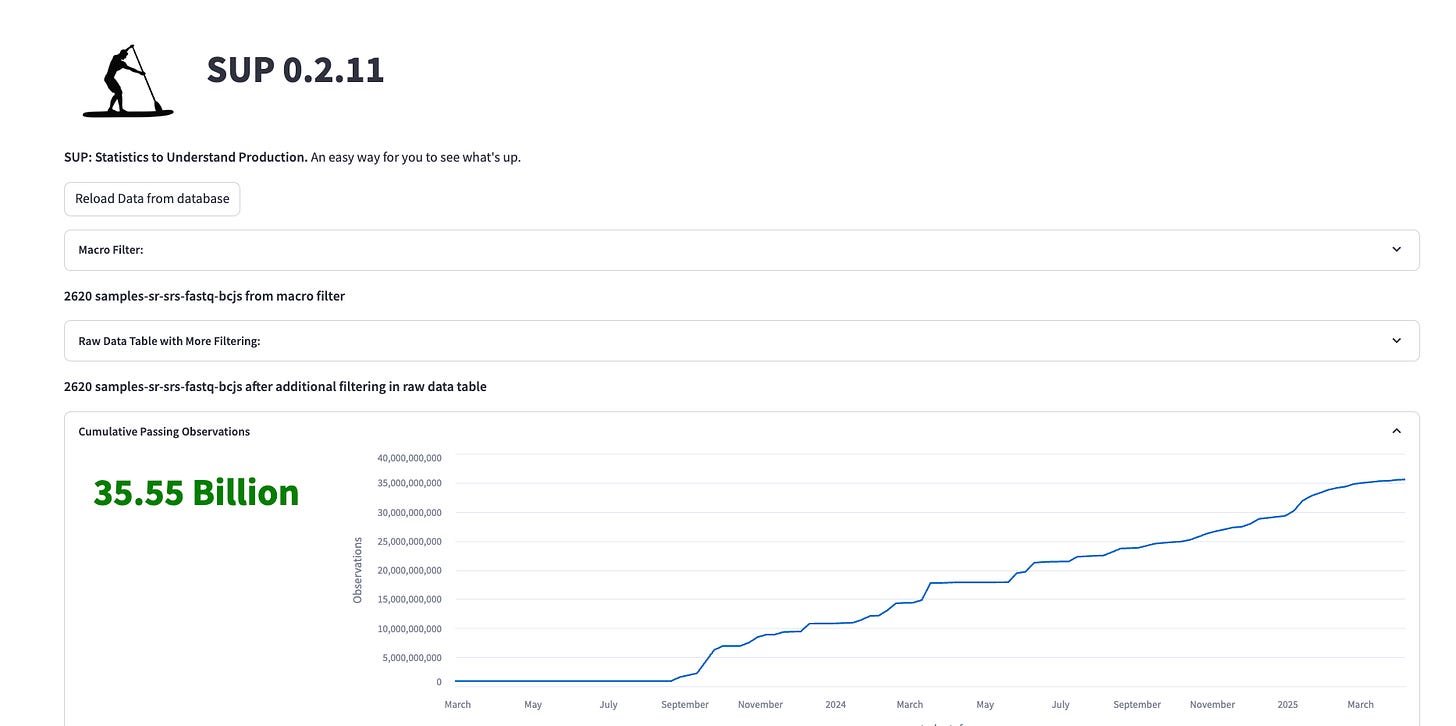
Over the past year we’ve continuously added capabilities to the lab. We’re now running everything from insect cell culture to NGS sequencing in house and have successfully screened over 380 different protein targets against millions of compounds in multiple conditions with a team of 3. I’ve been learning Python and polishing my dad-jokes. Here are a couple recent screenshots from our Statistics to Understand Production (SUP) dashboard:
Figure 1. We’ve been building Streamlit web-apps on top of SQLAlchemy databases with BigQuery to define and track all of our experiments. This investment is really starting to pay dividends as we scale up. We all use this SUP dashboard to see “what’s up” with our data generation and quality.
We’ve implemented a variety of critical instrumentation as well; a Hamilton Star liquid handler, a Tecan M200 plate reader, a much bigger Element Aviti sequencer, 2D barcode scanners, Kingfisher mag-bead separators, and more, mainly relying on used and auction sites to find good robotics from bad homes. This physical progress has been satisfying, and is a critical component to scaling our lab out, but I want to emphasize it’s not the most important thing.
Laboratory Automation Isn’t Really About Robotics
Since 2001, when I programmed my first PCR and protein purification methods on a Biomek-2000, I’ve helped scale and automate diverse biotechnology assays for a variety of companies. In that time, I’ve come to realize most are initially too focused on the physical aspects of automation. It’s the most visible aspect, but in most cases, the vast majority of the physical items that are handled by the automation are expensive to procure, used briefly, and then become hazardous waste that is difficult to safely discard. It is only the data that lives on. Whenever possible, these wasteful activities should be eliminated or simplified so that only the truly critical tasks are left to be automated. Focusing on process development can dramatically impact automation effectiveness, often making the difference between successful and unsuccessful implementation of expensive new instruments. Automating a baroque and complex process will not yield a good ROI.
Optimize Process First
Process optimization is one of the reasons I’m so excited to pursue DNA-encoded chemical libraries (DECL) at Leash. The DECL drug discovery workflow is a next generation technique that is heavily optimized. With previous classical high throughput screening (HTS) approaches, each molecule had to be measured individually. The DECL workflow uniquely tags small molecules as they are synthesized so that we can measure millions of molecules interacting with a drug target in a single well. By radically re-inventing the drug discovery screening process, the DECL workflow drastically reduces the need for physical automation and allows us to focus its application where it’s most critical: data generation. At Leash we’ve generated billions of measurements screening millions of compounds against hundreds of unique protein targets. These massive datasets are fueling our machine learning models and drug discovery programs, allowing us to pursue novel small molecule therapies for important human diseases.
Figure 2. Our investment in process development and automation has paid dividends as we scale the quantity, quality, and variety of different protein targets screened
Stay tuned for part 2 where I’ll cover our first steps: automating data collection.