Hit expansion at scale with Hermes
Fast binding prediction enables massive exploration of nearby chemical neighborhoods
TL;DR
Hermes is a compact, fast binding prediction model trained entirely on in-house DEL data collected here at Leash. Building on Hermes, we created Artemis, a hit expansion tool that applies chemist-inspired SMARTS transformations and uses a beam search–like strategy to explore chemical space efficiently. Hermes’s small footprint makes Artemis highly scalable: it runs reliably on low-cost cloud GPUs with SkyPilot’s bleeding-edge SkyServe features, keeping compute infrastructure under $5/hr while enabling thousands of candidate molecules to be evaluated in parallel. Beyond design, Artemis also acts as an explainability layer, revealing Hermes’s biases and feeding insights back into DEL screen design. Together, Hermes and Artemis form a closed-loop drug discovery pipeline — high-quality data fuels accurate models, models guide scalable exploration, and exploration uncovers new data needs.

In July, we released a blog post about Hermes (ref 1), our small molecule–protein binding prediction model trained exclusively on in-house data generated by our DNA-encoded library (DEL) platform. Hermes demonstrated three important points:
DEL screens, when designed carefully, provide a scalable and relevant training source for the binding prediction problem.
Hermes was trained only on Leash’s internal dataset, avoiding the noise, leakage, and batch effects common in public binding datasets.
Despite being a fraction of the size of other models, Hermes is 200–500x faster and shows superior performance to Boltz-2 on Leash’s own validation sets, with only a narrow performance gap on curated third-party benchmarks.
This performance validates Leash’s mission: data quality and scale combined can train models to make powerful out-of-distribution predictions. Our investment in generating binding measurements under tightly controlled conditions has paid off; Hermes generalizes well without requiring massive model complexity. Still, chemical space is vast and efficiently exploring this space is a difficult challenge.
Now, with Hermes as the engine, we’ve built Artemis, an internal hit expansion tool. Artemis leverages Hermes to explore chemical space around a target of interest, proposing thousands of plausible binders in minutes. Because Hermes is lightweight and efficient, Artemis runs on SkyPilot’s latest SkyServe features, which orchestrate inference endpoints across low-cost spot GPU instances. This allows Artemis to utilize its multi-GPU inference pipeline at <$5/hr.
Artemis
To use Artemis, a scientist provides two inputs: an amino acid sequence for the protein target and a one or more SMILES string representing starting small molecule(s). These are the same input types used by Hermes. Artemis then applies a curated library of SMARTS transformations, a type of rule-based chemical edits, to each SMILES input. This process is designed to mimic the intuition of a medicinal chemist exploring scaffold modifications, similar to the classic Topliss tree approach (Fig 1).
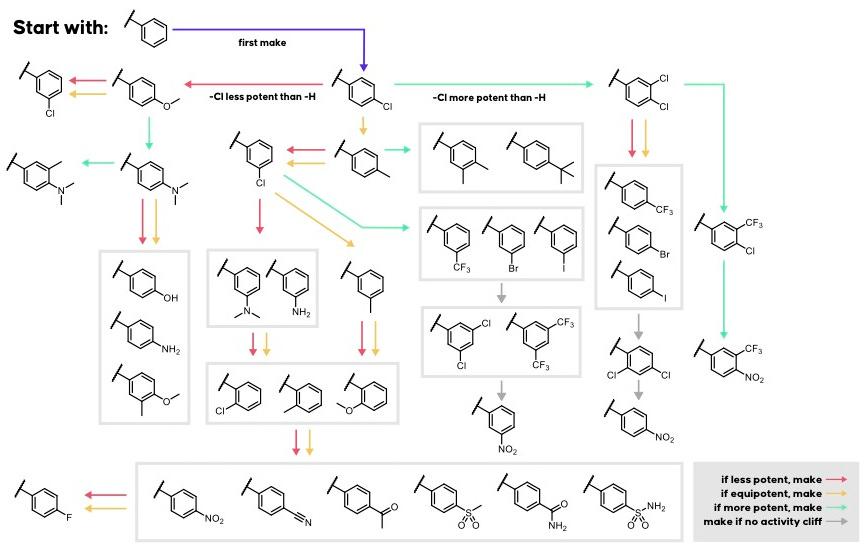
The user enters a protein target (amino acid sequence, UniRef ID, HUGO symbol) and a SMILES starting point (Fig 2). Each SMILES string becomes the root node of a search. Artemis applies its list of SMARTS reactions (such as those shown in Fig 1), and if the product of a reaction is chemically valid, it is added as a candidate for evaluation. For example, the transformation
[c:1][NH2:2]>>[c:1][NH:2]S(=O)(=O)C
converts an aniline into a sulfonamide group, a common substitution used to adjust polarity and binding properties.
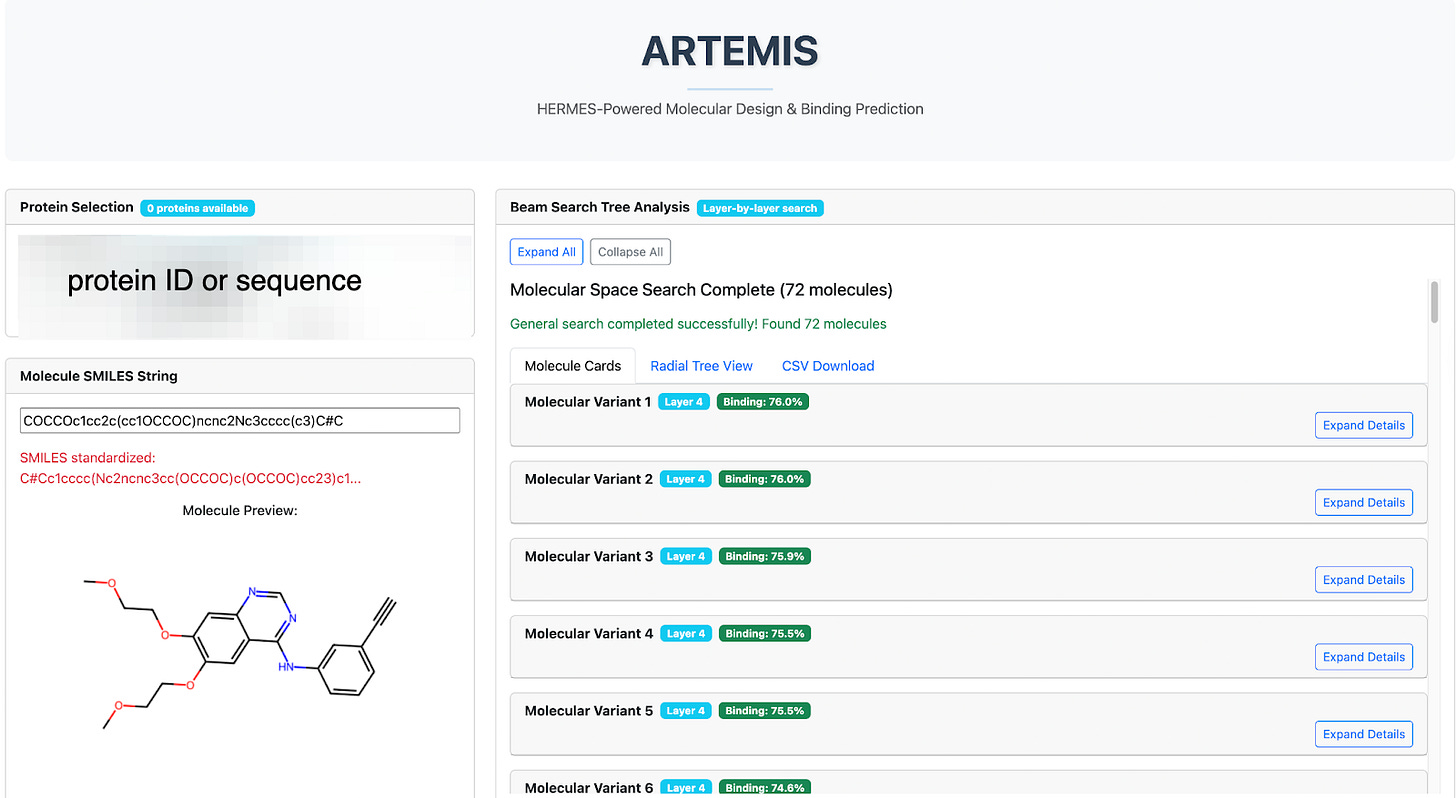
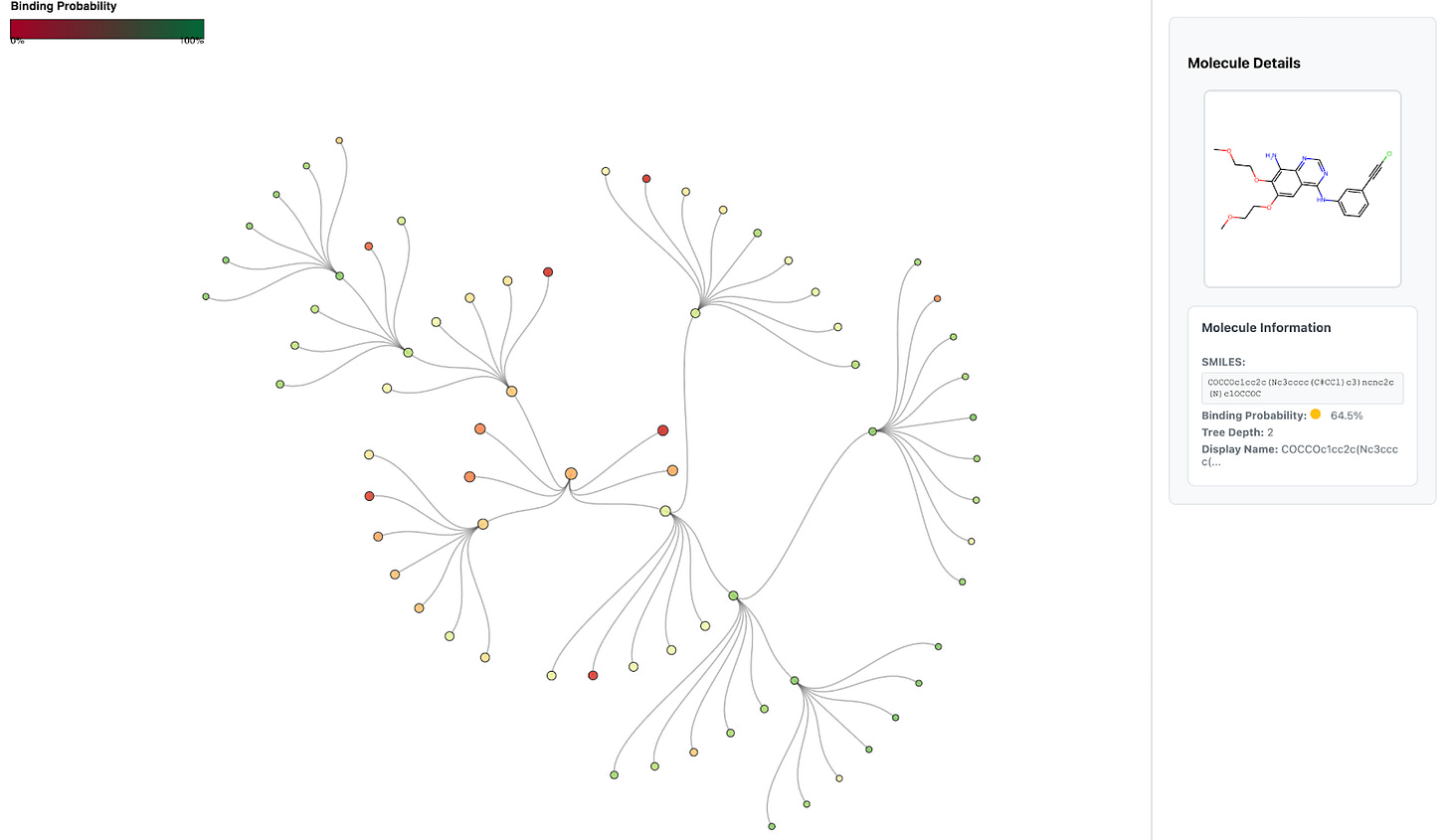
The resulting candidates form a layer of the search tree, which is batched and sent to Hermes for binding prediction. Once Hermes scores these molecules, Artemis selects the top-k most promising candidates (e.g., k = 3) to expand into the next layer (Fig. 3). This iterative pruning-and-expansion process is directly analogous to a beam search in machine learning: rather than exploring every possible modification, Artemis maintains a focused “beam” of the most promising molecules at each step.
For chemists, this means Artemis is systematically exploring chemical space in a way that mirrors human-driven hit expansion, but guided by Hermes’s predictions. For ML practitioners, the analogy to beam search highlights how Artemis balances exploration and efficiency when navigating a branching search space.
While Artemis currently uses a straightforward top-k expansion strategy for clarity and efficiency, the framework is flexible. For example, one could swap in a priority queue ranked by binding score or add diversity constraints to encourage exploration across different scaffolds.
Scaling
Expanding three molecules per layer, limiting the depth to three, and having a small collection of possible SMARTS reactions is great for local Hermes runs on a MacBook Pro M4 Max. But the true advantage of a model like Hermes isn’t that it can run on a laptop; it’s that this small footprint allows it to scale tremendously.
At scale, Artemis can pair Hermes with an arsenal of SMARTS reactions to explore much deeper corners of molecular space for large-scale campaigns. This unlocks broad discovery: thousands of diverse chemical variants can be evaluated computationally and then handed off to medicinal chemists for follow-up. Instead of screening one scaffold at a time, Hermes enables us to expand multiple scaffolds in parallel while still staying within modest computational budgets.
At Leash, we orchestrate this scaling using SkyPilot’s pre-release nightly build of SkyServe. This build allows us to deploy Hermes endpoints onto spot GPU instances across cloud regions, effectively renting leftover GPUs at steep discounts, while automatically handling preemptions. If a spot instance is reclaimed, SkyServe seamlessly redeploys Hermes so Artemis never stalls waiting for compute.
Because Hermes is lightweight, it fits comfortably on a single NVIDIA L4 instance on AWS. Across regions, these spot GPUs reliably cost <$0.75/hr. That low footprint means we can run multiple parallel Hermes endpoints for less than the price of a single high-end GPU used by larger models. SkyServe also lets us scale to zero when Artemis isn’t active, so Leash doesn’t pay for idle infrastructure.
The combination of a compact model, elastic GPU orchestration, and efficient cloud economics turns Hermes from a research demo into a production-grade engine for hit expansion. Scientists gain scale and speed, and we keep costs well below those of traditional computational chemistry approaches.
Explainable AI
Leash generates new protein–ligand binding data daily in our lab (on big days, ~100M measurements), and our data scientists curate that stream to retain the most informative measurements for training state-of-the-art binding prediction models. Hermes was trained on this curated dataset and benchmarked against the best models in the field. Now, Artemis not only leverages Hermes to search chemical space but also acts as a lens into Hermes’s decision-making.
Because Artemis expands and ranks molecules systematically, it provides a natural way to examine which chemical transformations Hermes favors or avoids. Patterns in these preferences help us spot biases in Hermes’s predictions, for example, tendencies toward certain scaffolds or underestimation of specific substitutions. These insights directly inform how we design new DEL screens and how we prioritize data collection, ensuring that Hermes (and future models) are continually exposed to the right challenges.
In this way, Artemis closes the loop: it is not just a generative tool for molecular design, but also a point of reference for explainability. By surfacing where Hermes succeeds and where it struggles, Artemis helps refine both our datasets and our models, bringing Leash’s drug discovery pipeline full circle. Please reach out if these ideas are of interest!
We thank the entire Leash team for data, insights, hustle, and grit.