This is the third in the continuing series:
PART 3
Automating Data Analysis:
Once it’s possible to automatically acquire hundreds or thousands of data points, analysis can become a bottleneck. What are the quality metrics of the laboratory? How often are standards compared? What is an acceptable range for each metric and how are you modeling its distribution? How do you define and track replicate consistency? Building systems that can efficiently summarize billions of data points is no small task. Defining tables, fields and calculations that can be automatically orchestrated to track this is not easy. It requires continual cross-functional discussion between scientists, business leaders, and developers. These metrics are important to decision making around what programs should be initiated, what optimizations should be pursued. Robust automation of data analysis creates a shared understanding across the organization, allowing for faster and more consistent decisions by all employees.
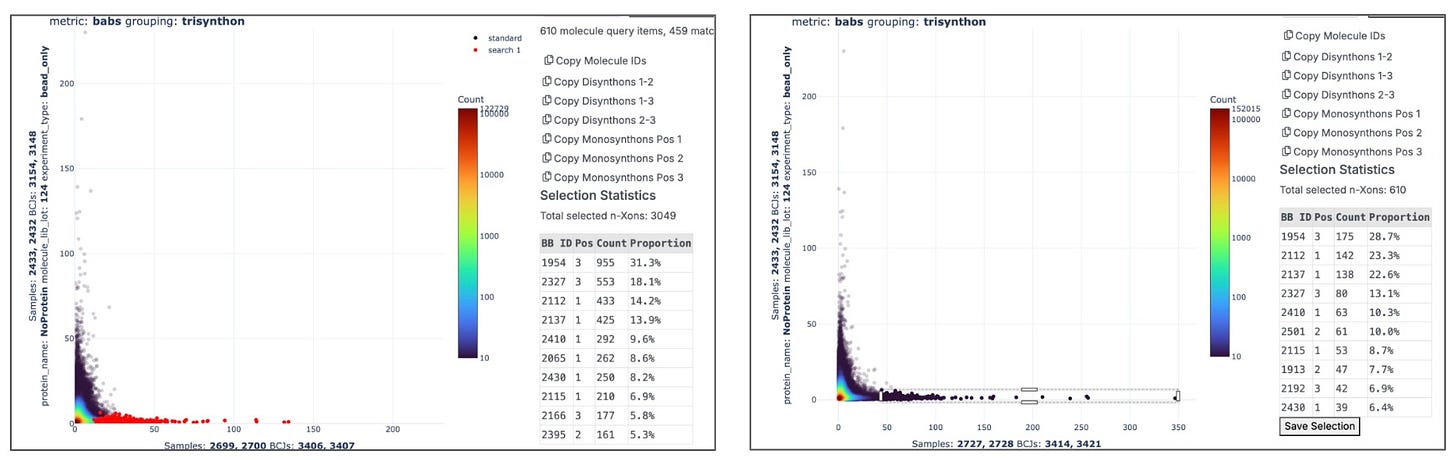
Our internal data review application Chimera displays two different protein constructs run at two different concentrations prefer the same set of 610 molecules out of 6.5M.
A screenshot showing the correlation between >120 different proteins’ selectivity for millions of molecules. The darker the cell, the more hits those two proteins share in common.
Automatically generating valid comparisons between replicates requires the consistent registration of metadata about each experiment we run. We’ve chosen to build this system in-house giving us the flexibility and control to quickly build new systems.
Automating the Scientific Method:
This concept of registration is critical when applied to experiments and their hypotheses. To rigorously apply the scientific method means to define your hypothesis before performing the experiment, and then to follow through to rigorously evaluate results against the hypothesis. In a fast moving company it can be difficult to ensure that this is done rigorously. At Leash, we realize that the scientific method is crucial to ensuring data-driven decision making. By focusing on data automation we save scientists time, and ensure consistently and rigorously analyze and review the datasets feeding our machine learning models as quickly as we can generate them.
Metadata about each selection is carefully collecting allowing us to automatically define valid groupings of experiments and their controls.
Experiments Have a Lifecycle:
Experiments are the unit of work here at Leash. Each time we explore a combination of molecules and drug target we register a new experiment with a defined hypothesis. We track that experiment through a lifecycle from creation to quality review. This is a critical aspect of the scientific method that is infrequently automated, perhaps because the process development work is under-valued and changing too rapidly. Defining a unit of work and tracking its manufacturing process is critical to reproducible science. This can be difficult and nuanced to do and will evolve over the life of the laboratory, but investing in it early has paid significant dividends here at Leash.
When an experiment is registered in our internal systems it streamlines decision making and data processing in the lab. Our web applications display available proteins, libraries and primers to assign to each sample. As these entities are assigned automated data analysis pipelines can be used to process pools of samples in a single NGS run. We take in the XML file from the plate reader and automatically assign protein and DNA concentrations to the database, which allows for downloading of automated pipetting instructions. Quality control dashboards allow for the selection of replicates based on experimental metadata and all of these data automations allow our scientists to quickly build and analyze massive datasets at scale. This creates the critical guardrails needed to monitor the performance of an automated laboratory and ensures the lab is ready to scale.
This careful and consistent focus on process development, data automation, quality metrics, and finally physical automation is what has allowed Leash to scale from 3 proteins and 3.2B measurements last year to 447 proteins and 41B measurements today. These datasets are fueling our machine learning and drug discovery programs ultimately helping to bring new therapeutics to patients.
With input and output data captured, internal data acquisition automated, and quality control methods in place, it is finally time to automate physical work in the lab and deploy robotics.
Stay tuned for part 4 where I’ll finally talk about robotics a little.






what is the computational burden from an analysis of this size? How long does it take, and on what type of hardware?